Grad Student Develops Program to Increase Accuracy of Community Formation Processes
November 22, 2019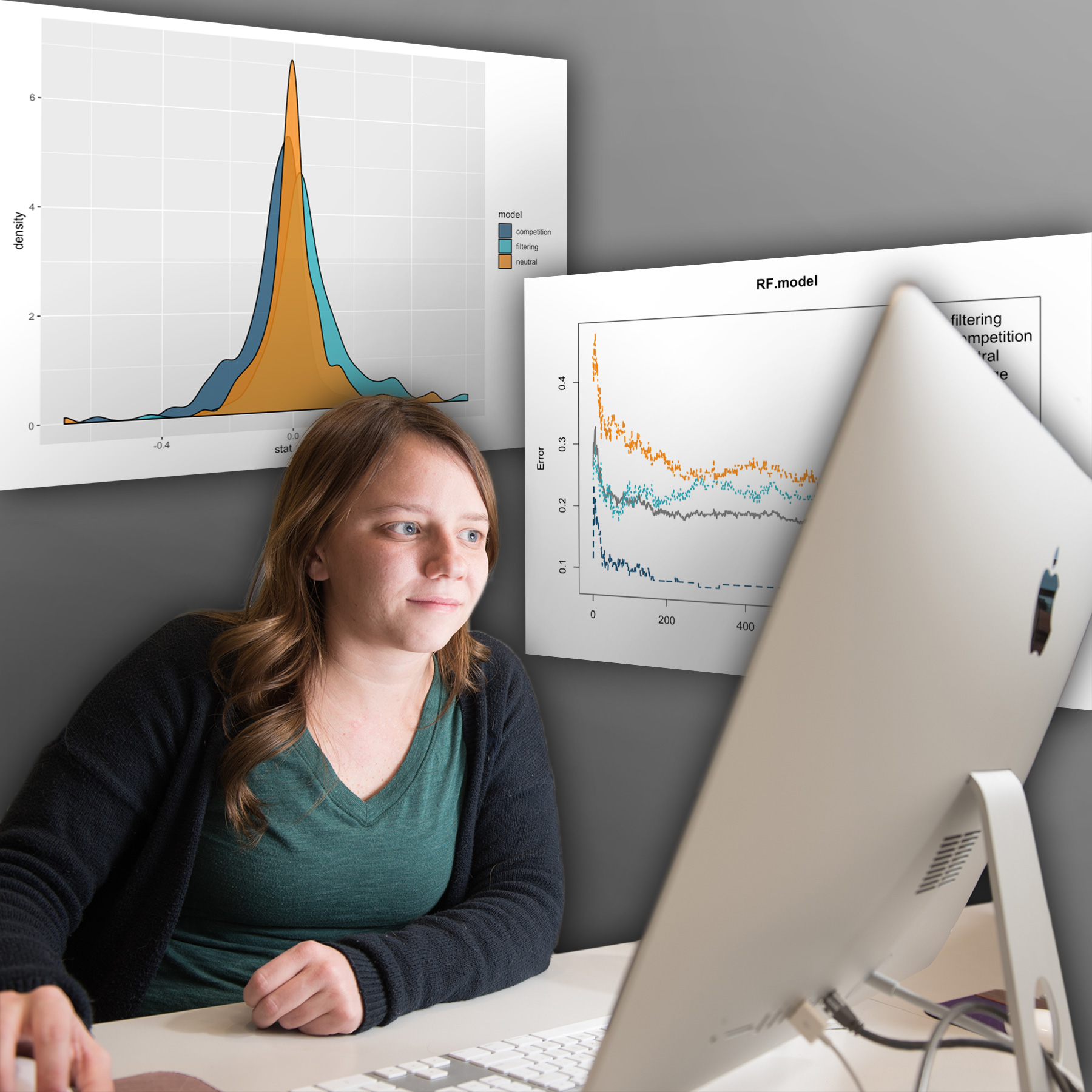
Ph.D. candidates Megan Ruffley, Katie Peterson, and Bob Week recently had a paper published in Ecology and Evolution titled “Identifying Models of Trait-Mediated Community Assembly Using Random Forests and Approximate Bayesian Computationâ€. Ruffley and Week are part of the Bioinformatics and Computational Biology graduate program, and Peterson comes from the Biology graduate program. Professors Dave Tank, Jack Sullivan, Scott Nuismer, and Christine Parent from the Biological Sciences Department were also all involved as the students’ mentors.
This publication is the second chapter of Ruffley’s dissertation and the culmination of her lab rotation in Luke Harmon’s lab. Ruffley designed a program called Community Assembly Model Inference, or CAMI. It uses a stochastic algorithm to simulate communities assembled under environmental filtering, competitive exclusion, and neutral species assembly processes—simultaneously considering phylogenetic and phenotypic information from species in local and regional communities. During the early stages of CAMI’s development, Bob Week provided key insights into the process of formally modeling the interactions among species within a community. These models then ultimately laid the foundation for the non-neutral community assembly processes implemented in CAMI. Collaborator Katie Peterson provided the first empirical dataset that CAMI was implemented on.
The paper and the creation of the accompanying CAMI program involved students and faculty with a wide range of research areas and skill sets. “The whole project was really a nice, big UI collaborationâ€, said Ruffley. The cost of the publication in Ecology and Evolution—an open access journal—will be covered by the university’s Open Access Publishing Fund.
Article by Katy Riendeau
IBEST Design & Marketing Coordinator